
Such optimization is necessary to enable the full development of artificially expanded genetic systems utilizing an expanded genetic code, thereby allowing for the site-specific incorporation of novel functional components (such as unnatural amino acids) into proteins. (b) qPCR and artificially expanded genetic systems: A number of research groups have been working on optimizing PCR amplification on templates containing 5-Me-iso-dC. The limits of detection of the assay were improved 10-fold, from < 500 HIV molecules/mL to < 50 molecules/mL. Use of this strategy resulted in a significant reduction in non-specific hybridization of the above three sequence types to non-target nucleic acid sequences, and thus less amplification of background. For example, Collins and co-workers significantly improved the sensitivity of a branched DNA quantitative hybridization assay for detecting the HIV POL sequence by incorporating ~30% 5-Me-iso-dC and iso-dG into the pre-amplifier, branched DNA (bDNA) amplifier and alkaline phosphate probe sequences used in the assay (3). (a) Molecular recognition: The 5-Me-iso-dC:iso-dG base pair has been incorporated into hybridization assays to enhance probe-target specificity and reduce spurious hybridization to non-target sequences. The combination of 5-Me-iso-dC’s high selectivity for iso-dG, and the resulting base pair’s high thermodynamic stability, make this modified base particular attractive in the following applications:
KLENOW FRAGMENT MOLECULAR WEIGHT ISO
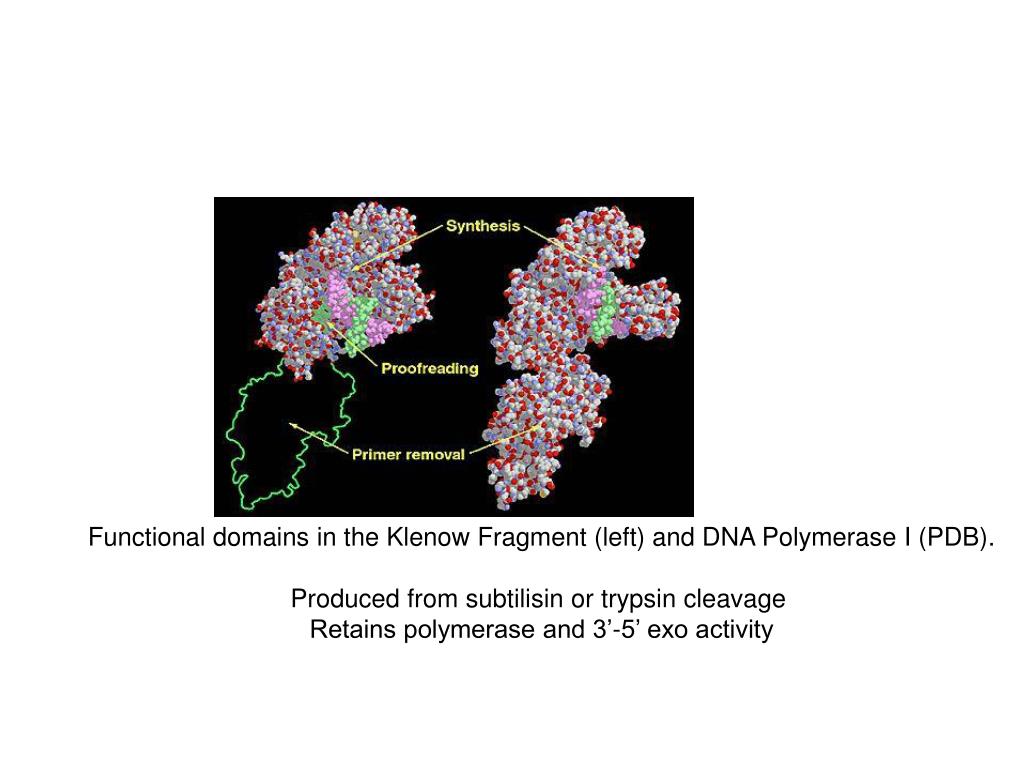
Furthermore, since iso dC and 5-Me-iso-dC does not pair with dG, iso dC and 5-Me-iso-dC:iso-dG can function as a stable unnatural base pair that can be used to expand the genetic code. Substitution of a 5-me iso-dC:iso-dG base pair for a C:G pair increases the Tm of the resulting duplex by ~2degC per base pair substitution (1,2). Ollis, D.L., Brick, P., Hamlin, R., Xuong, N.G., Steitz, T.A.ĭepartment of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511.Iso dC and 5-methyl iso-deoxycytosine (5-Me-iso-dC) forms a Watson-Crick base pair with iso-dG, but has a different type of hydrogen bonding pattern than those observed for the natural base pairs A:T and C:G. Structure of Large Fragment of Escherichia Coli DNA Polymerase I Complexed with dTMP.Co-Crystal Structure of an Editing Complex of Klenow Fragment with DNAįreemont, P.S., Friedman, J.M., Beese, L.S., Steitz, T.A.Structural Basis for the 3'-5' Exonuclease Activity of Escherichia Coli DNA Polymerase I: A Two Metal Ion Mechanism.Although this cocrystal structure appears to be an editing complex, it suggests that the primer strand approaches the catalytic site of the polymerase from the direction of the 3' to 5' exonuclease domain and that the duplex DNA product may bend to enter the cleft that contains the polymerase catalytic site. A single-stranded, 3' extension of three nucleotides bound to the 3' to 5' exonuclease active site. When the fragment bound DNA, a region previously referred to as the "disordered domain" became more ordered and moved along with two helices toward the 3' to 5' exonuclease domain to form the binding groove. Klenow fragment of Escherichia coli DNA polymerase I, which was cocrystallized with duplex DNA, positioned 11 base pairs of DNA in a groove that lies at right angles to the cleft that contains the polymerase active site and is adjacent to the 3' to 5' exonuclease domain. Diversity, Equity, Inclusion, and Access.Citation, Usage, Privacy Policies, Logo.Biologically Interesting Molecule Reference Dictionary (BIRD).


 0 kommentar(er)
0 kommentar(er)
